Gene Set Correlation Enrichment Analysis
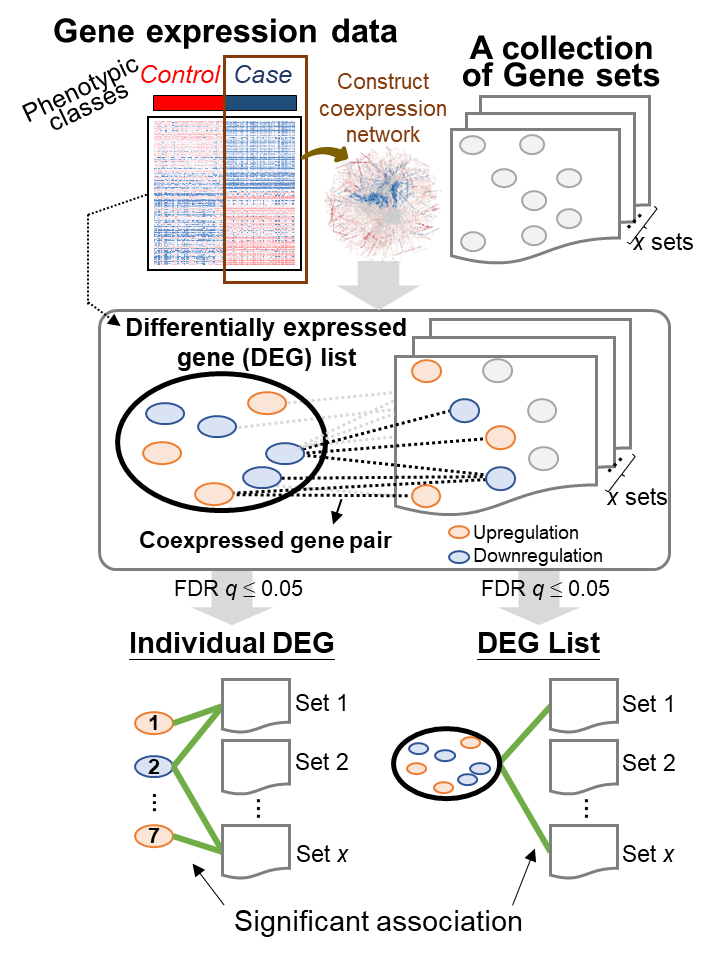
Gene Set Correlation Enrichment (Gscore) is an online tool for interpreting and annotating gene expression dataset by using a dataset-derived coexpression network to measure the statistical significance of associations between your interesting differentially expressed gene (DEG) list (i.e., query) and the collection of gene sets. Based on the hypergeometric distribution with Benjamini–Hochberg correction, the tool identifies the coexpressed gene pairs between:
(1) individual DEG in the query DEG list and all the DEGs of a certain gene set in the selected collection to determine its association significance;
(2) query DEG list and all the DEGs of a certain gene set in the selected collection to determine its association significance.
This online tool is free and open to all users and there is no login requirement.
Bring your own data / Select your interesting data:
(1) individual DEG in the query DEG list and all the DEGs of a certain gene set in the selected collection to determine its association significance;
(2) query DEG list and all the DEGs of a certain gene set in the selected collection to determine its association significance.
This online tool is free and open to all users and there is no login requirement.
Bring your own data / Select your interesting data:
Upload your genome-wide expression profiles with samples belonging to two classes (i.e., cases and controls) or select one of the disease-related expression profiles collected from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases.
Upload your own gene sets or select one collection of annotated gene sets collected from several public databases, such as Kyoto Encyclopedia of Genes and Genome (KEGG) database, Gene Ontology (GO), and Molecular Signatures Database (MSigDB).